Developmental Biology Research
at Illinois Wesleyan University

Characiform fishes are a large assemblage of tropical fishes which inhabit the freshwaters of North America, South America, and Africa. They are considered to be closely related to siluriforms (catfishes) and cypriniforms (minnows) due to their shared modification of anterior vertebrae to facilitate auditory reception (the Weberian apparatus). Over 1600 species of characiform fishes have been described, and together, they exhibit a remarkable range of size, shape, and color. However, little is known about their life histories, including embryology and larval growth.
My interests lie in examining the ontogeny of these fishes to uncover the developmental mechanisms responsible for the skeletal variations seen in the adults. This involves the analysis of their development at the organismal, cellular, and molecular levels.
There are a number of aspects to my research program:
Analysis of characiform embryology and larval development.
Very little is known regarding the development of these fishes. This aspect of my research deals with the spawning of adults and the analysis of both embryonic and larval development. By comparing the development of different species, the differences leading to morphological variations can be ascertained, prompting further investigations.
Comparison of characiform development with that of other fishes.
Danio rerio is a cypriniform fish prevalent in developmental biology research, and represents a logical point of comparison. Differences in developmental morphologies between D. rerio and characiform fishes may serve as a basis for further investigations, providing insight into mechanisms of evolutionary change.
Cloning and analysis of developmental genes.
While morphological and embryological analyses can provide particular insights in regard to developmental variation, molecular-based research can provide a more in-depth understanding of how variation is achieved. Danio rerio represents a rich source of molecular data, and the knowledge gained from D. rerio can be translated to studies of characiform fishes. Much of this research involves purifying specific gene products (i.e., mRNAs) of interest from embryos, cloning those mRNAs, and performing analyses to define when and where the mRNAs are expressed in the embryo. Differences between the expression of the genes will likely reflect differences in morphologies between species.
Current projects
Larval development of the characiform fish Moenkhausia sanctaefilomenae. Numerous developmental changes occur throughout the larval period in fishes, and these changes directly influence the final morphology of the adult. I am currently examining the patterns of allometric growth and pigmentation in the larvae of M. sanctaefilomenae in order to ascertain the time frames of and magnitude of these phenomena.
Growth and shaping of jaw elements in Moenkhausia sanctaefilomenae. The examinations of early skeletal development in M. sanctaefilomenae have revealed a great degree of growth in the jaw elements of early larva. The cellular mechanisms producing this growth are being explored using markers for proliferation.
Cloning and expression analysis of genes with roles in jaw development. The skeletal elements of the jaw come from a group of cells known as cranial neural crest (CNC) cells, and these cells form cartilage and bone under the direction of surrounding tissues. By examining the expression of mRNAs made by CNC cells and surrounding tissues within both M sanctaefilomenae and D. rerio, we can account for the observable morphological differences between both species. Some current genes currently under investigation include Pax1, Pax9, Sox9, and Foxd3.
Publications
Walter B.E. Early ontogeny of aquarium-raised Moenkhausia sanctaefilomenae (Characiformes: Characidae). (submitted)
Walter B.E. Cranial skeletogenesis and osteology of the redeye tetra Moenkhausia sanctaefilomenae. (submitted)
Walter B.E., Perry K.J., Fukui L., Malloch E.L., Wever J., Henry J.J. Psf2 plays important roles in normal eye development in Xenopus laevis. Molecular Vision. 2008 May 19; 14:906-21.
Walter B.E. and Henry J.J. Embryonic expression of pre-initiation DNA replication factors in Xenopus laevis. Gene Expression Patterns. 2004 Nov; 5(1):81-9.
Walter B.E., Tian Y., Garlisch A.K., Carinato M.E., Elkins M.B., Wolfe A.D., Schaefer J.J., Perry K.J., and Henry J.J. Molecular profiling: gene expression reveals discrete phases of lens induction and development in Xenopus laevis. Molecular Vision. 2004 Mar 24; 10:186-98.
Henry J.J., Carinato M.E., Schaefer J.J., Wolfe A.D., Walter B.E., Perry K.J., and Elbl T.N. Characterizing gene expression during lens formation in Xenopus laevis: evaluating the model for embryonic lens induction. Developmental Dynamics. 2002 Jun; 224(2):168-85.
Carinato M.E., Walter B.E., and Henry J.J. Xenopus laevis gelatinase B (Xmmp-9): development, regeneration, and wound healing. Developmental Dynamics. 2000 Apr; 217(4):377-87.
Embryology of characiform fishes and
developmental mechanisms of skeletogenic variation


Gasteropelecus
Hemigrammus


Moenkhausia
Nannostomus

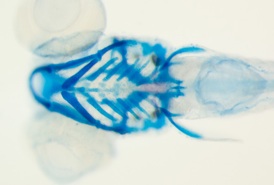
comparison of similarly-aged larvae between Danio rerio and glowlight tetra Hemigrammus erythrozonus reveals marked disparities in the size and shapes of the skeletal elements

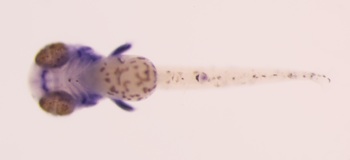
Pax9
Sox9
Danio
Hemigrammus
expressions of Pax9 and Sox9 in the embryos of Moenkhausia sanctaefilomenae